You are here: Urology Textbook > Urologic examinations > Urine culture
Urine Culture: Colony Count and Antibiotic Resistance
- Urine Analysis: sediment and dipstick
- Urine Analysis: urine culture
- Urine Analysis: 24-hour urine test
Preanalytic Phase
Indications for urine culture in asymptomatic patients
- Leucocyturia, hematuria, or positive nitrite test in patients with risk factors (immune suppression, vesicoureteral reflux, bladder function disorders).
- Pregnancy (screening for asymptomatic bacteriuria).
- Before and after surgery of the urinary tract.
- After treatment of pyelonephritis or complicated UTI.
Indications for urine culture in symptomatic patients:
- All patients with UTI, not mandatory in women with uncomplicated UTI.
- Unclear abdominal pain or flank pain.
- Before treatment of recurrent UTI.
- Before treatment of suspected nosocomial UTI.
- All patients with fever or sepsis.
Collection of specimen
The risk of bacterial contamination is as high as 40% with noninvasive urine collection, depending on sex, age, and compliance. The first choice for urine collection is sterile bladder catheterizations, during interventions or by suprapubic bladder puncture. Otherwise, urine collection is possible by a midstream clean-catch specimen or with an adhesive bag in infants.
Four-glass test:
The four-glass test is used to identify chronic bacterial prostatitis. Four specimens are collected and examined microbiologically with urine sediment microscopy and culture. Bacterial prostatitis is likely if there is a 10-fold increase in bacteria between VB1/2 and EPS/VB3:
- The first 10 ml of urine (urethral specimen, VB1)
- Midstream urine (bladder specimen, VB2)
- Expressed prostate secret (EPS): standard values are <10 leucocytes per view in high magnification.
- The first 10 ml of urine after prostatic massage (prostate specimen VB3)
Due to the high costs and effort, a two-glass test has become standard, and recent studies have confirmed the equivalence:
- A midstream urine (bladder specimen)
- The first 10 ml of urine after prostatic massage (prostate specimen)
Transport of the specimen:
The examination of fresh urine within two hours is best. A cooled specimen or a urine culture transport tube (containing boric acid) should be used for longer transport.
Quantitative Urine Cultur: the Meaning of the Colony Count (cfu/ml)
Quantitative urine culture determines the colony count of a urine specimen. The most commonly used technique uses standardized plastic loops to distribute 10 μl urine on an agar plate (e.g., CLED-Agar) [fig. technique of quantitative urine culture]. The plates are incubated for 24 h at 36 degrees Celsius. One colony equals 102 colony forming units per ml (cfu/ml), 10 colonies = 103 cfu/ml, 100 Kolonien = 104 cfu/ml, and 1000 colonies = 105 cfu/ml. The colony count is determined either with counting or with a semiquantitative technique by comparing the agar plate with standardized figures.
 |
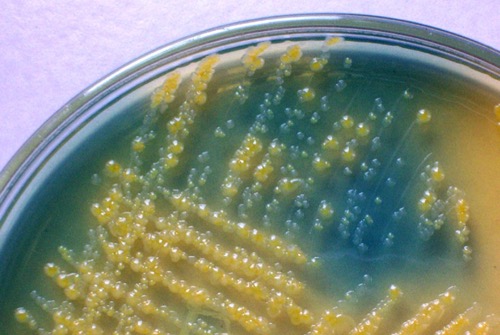
CLED-Agar:
CLED is a nonselective agar and used for quantitative urine culture. The agar enables the growth of all typical bacteria of Urinary tract infections. CLED stands for Cystine Lactose Electrolyte Deficient-Agar, the low concentration of electrolytes prevents swarming from proteus species. Bacteria with the fermentation of lactose will produce yellow colonies and stain the agar yellow. Bacteria without fermentation of lactose (non-fermenters) will build grey to blue colonies [fig. bacterial colonies on CLED agar].
 |
Significant bacteriuria:
≥ 105 colony-forming units in a midstream urine specimen speak in favor of significant bacteriuria (Kass et al., 2002). In case of high diuresis, frequency, or sterile urine specimen collection, even a lower bacteria count may represent a significant sign of Urinary tract infections.
Identification of Bacteria
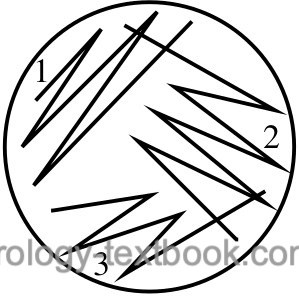
Identifying the bacteria makes sense if there is no mixed bacterial growth on the CLED agar plate (not more than two germs). Diverse bacterial growth suggests contamination during urine collection; the urine culture should be repeated with a new specimen. The correct identification of the bacteria will only be successful if isolated colonies are present on the agar plates. Mixed growth may remain unnoticed in confluent colonies. If in doubt, repeat streaking on a new agar plate to achieve isolated colonies [fig. streaking technique for the isolation of bacterial colonies].
 |
For quick identification, the following selective agar plates are inoculated parallel to quantitative urine culture (on CLED):
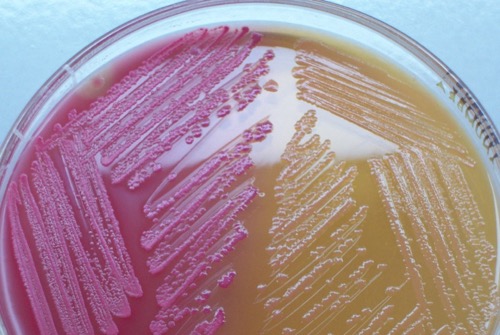
MacConkey agar:
MacConkey is a selective agar for the growth of Gram-negative bacteria. The fermentation of lactose leads to a pink color of the colonies [fig. bacterial colonies on MacConkey agar], non-fermenters produce colorless colonies.
 |
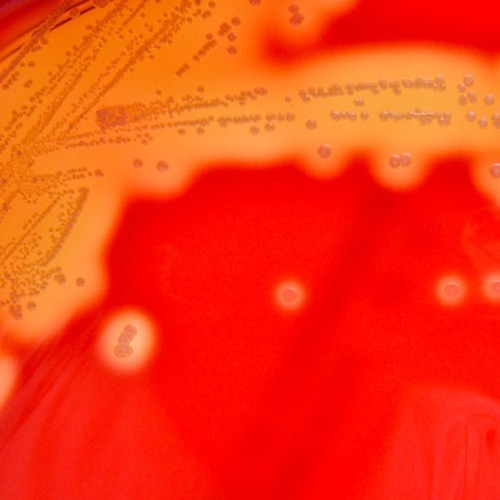
Columbia-CNA agar:
Columbia-CNA agar is a selective agar for the growth of Gram-positive bacteria (e.g., streptococcus, enterococcus, and staphylococcus). The agar contains the antibiotics colistin and nalidixic acid, which prevent the growth of Gram-negative bacteria. The agar contains intact erythrocytes. The metabolism of the bacteria causes different forms of hemolysis: α hemolysis (green color of the agar due to oxidation of hemoglobin), β hemolysis (transparent color of the agar due to complete hemolysis) and γ hemolysis (unchanged agar color) [fig. β hemolysis on Columbia CNA agar].
 |
Sabouraud-Dextrose agar:
Sabouraud-Dextrose agar is a selective agar for the growth of fungi like candida and aspergillus.
Approximate identification of bacteria:
Approximate identification of bacteria is possible with the help of the growth on above mentioned selective agar plates and basic biochemical properties:
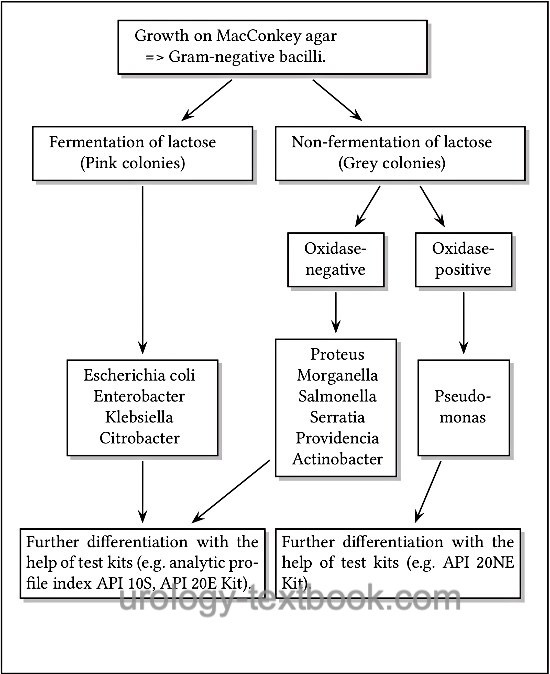
- Growth on MacConkey agar => Gram-negative bacillus. Further differentiation is possible with the help of lactose fermentation and the oxidase test [fig. approximate differentiation of Gram-negative bacteria].
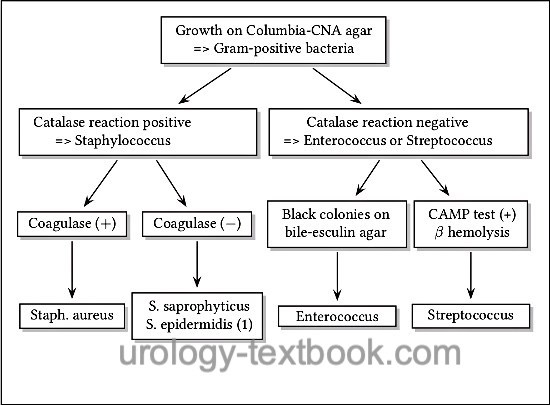
- Growth on Columbia-CNA agar => Gram-positive bacteria. Further differentiation is possible with the help of the catalase test, hemolysis, growth on bile-esculin agar, CAMP test, and sensibility to novobiocin [fig. approximate differentiation of Gram-positive bacteria].
- Growth on Sabouraud-Dextrose agar => fungi like candida.
 |
 |
Exact identification of bacteria:
The exact identification is possible with the help of test kits using biochemical properties of the bacteria: e.g., oxidase, catalase, laktase, fermentation, urease, mobility, and usage of citrate. Examples of test kits are the API (Analytic Profile Index) or VITEK system from the company bioMérieux or the RAS-ID system from the company BIO-RAD.
Antibiotic Sensitivity Test
A defined amount of individual colonies are inoculated on a Müller-Hinton agar plate. Paper plates with different concentrations of antibiotics are placed on the agar plate. The antibiotic will diffuse into the agar and inhibit bacterial growth depending on sensitivity. After incubation of 24 h, the diameter around the paper plates is suggestive of the minimum inhibitory concentration for the tested antibiotic. The minimum inhibitory concentration (MIC) is the lowest concentration of an antibiotic which prevents visible growth of a bacterium.
EUCAST:
Recommendations for the technique of antibiotic sensitivity tests are published, e.g., in Europe from EUCAST (European Committee on Antimicrobial Susceptibility Testing). Details on the homepage https://www.eucast.org. Several commercial systems simplify the antibiotic sensitivity test, e.g., RAS-ID from the company BIO-RAD or VITEK system from the company bioMérieux.
| Urine analysis | Index | 24-hour urine |
Index: 1–9 A B C D E F G H I J K L M N O P Q R S T U V W X Y Z
References
 Deutsche Version: Urinkultur
Deutsche Version: Urinkultur